Effect of Storage on the Quantity and Quality of DNA Isolated from Preserved and Discarded Blood: Forensic Applications
| Received 27 Apr, 2020 |
Accepted 13 Aug, 2020 |
Published 01 Jan, 2022 |
Background and Objective: The preservation of DNA is of eclectic attention to scientists in different fields of studies. The target of this study was to investigate if storage has any effect on the quantity and quality of DNA isolated from blood. Materials and Methods: A total of ten patients were used for the study and 20 blood samples were collected and fashioned into two precise categories, freshly preserved and carelessly discarded blood samples. Each of the categories contained 4 females and 6 males. The preserved and discarded groups were kept in the EDTA bottle at 4°C and in the syringe kept in dump site, respectively. DNA was extracted using phenol-chloroform method while the concentration and quality of DNA was measured spectrophotometrically and electrophoretically, respectively. Results: The concentrations of DNA in the preserved blood samples showed a significant increase compared to discarded samples, t (18) = 5.96, p = 0.00. Also, there was a significant increase in the quality of DNA obtained from the preserved blood samples when compared to the discarded groups, t (18) = 9.97, p = 0.00. Results of gel electrophoresis indicated that DNA from preserved blood was more intact and most probably had higher molecular weight and less shearing than the DNA compared to discarded blood. Conclusion: The results of the study showed that preservation has serious effects on the quantity and quality of DNA extracted. However, both samples are good for forensic investigations and molecular diagnosis of genetic diseases but preserved is better of.
| Copyright © 2022 Nwankwo et al. This is an open-access article distributed under the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited. |
INTRODUCTION
Whole blood is a frequent foundation of DNA, mainly for scientific, forensic, pharmaceutical and genotype diagnostic facilities. Nowadays, blood samples are collected in quite a lot of research laboratories via mail and/or preserve the blood samples proceeding to DNA mining1. Specific circumstance primarily observed in forensic sciences is that blood samples are most often inadequately preserved or even abandonedbefore the extraction of DNA from them is required. Therefore, the effects of storage capacity and time as well as storage situation and heat on DNA produced are of great concern2,3.
There is a growing curiosity in the conservation of DNA from many fields of studies viz., medical, pharmaceutical, forensics science, homeland defense, molecular biology and bio-repository management etc. Although, each of these fields is concerned with the maintenance of DNA integrity, the requirements in terms of storage time and fidelity4,5. When discussing strategies for preservation, both storage time and fidelity must be vigilantly considered and defined.
Current sequencing technology depends on the Polymerase Chain Reaction (PCR) to make copies of stored DNA that are ultimately used for sequence identification. As a result of this process, accurate sequence information can be obtained even from samples in which the DNA has been reduced to fragments. Although, the desire for sequence identification allows substantially greater levels of degradation to be tolerated, bio-repositories typically strive to preserve samples permanently so that future scientists can utilize specimens for studies at some undefined point in time4,6.
Some restricted degradation occurs in sample regardless of storage conditions4,7 and the goal of preservation is to minimize this rate of degradation. In most instances, DNA samples are stored at -80°C or in liquid nitrogen (-196°C) but there is a significant expense associated with maintaining these conditions in samples8. Previous studies of DNA in solution have clearly shown that degradation rates can be precisely predicted and buffer conditions can be adjusted to obtain remarkable stability at room temperature4,9.
Long term storage (LTS) of whole blood assortment can be a very important resource of DNA with no gathering costs but there is a paucity of evidence on techniques effective to extricate genomic DNA from such type of biological substance2. Genomic DNA obtained from entire blood is a precious resource. Preferably, abstraction can take place instantly after assemblage of the blood model but this is not always feasible due to reasons emanating from logistics and finances. This simply implies that total-blood samples may go through prolonged storage for months or even years before handling. Insufficient storage capacity, especially in the case of long-term storage can adversely influence DNA extraction and quality which can lead to misuse or imprecise investigational findings1.
Bulla et al.10 examined the yield and quality of genomic DNA extracted from whole blood after prolonged storage at separate high temperature, both with and without a stabilizing mediator. The results proved that the conditions of storage have a momentous impact on the extraction and yield of DNA but little or no effect on DNA integrity and methylation. It also showed that the storage of EDTA blood at -80°C guarantees high-quality DNA with a good yield. It is a common experience that the premium way to appropriate DNA is to use crisp material as the foundation of raw material. In several circumstances, new tissue cannot be achieved and one has to rely on abandoned or previously accumulated and stored samples. When blood is the resource tissue, it is often refrigerated instantly and stored at low temperatures. The isolated capacity of DNA from whole blood hangs upon the feasibility of the leukocytes. The feasibility of granulocyte11 in whole blood reduced for about 14% in 24 hrs at 4°C and numerous surveys by McCullough et al.11 and Glasser et al.12 recommended 22°C as optimal temperature for storage of granulocytes. However, it has remained that low yields of DNA may result, notwithstanding the high white blood cell counts, because of the use of inadequate mining solution13. The type and intensity of anticoagulant used or other additives may affect DNA extraction.
The current study was undertaken to compare the quantity, quality and stability of DNA extracted from EDTA preserved blood samples and stored at 4°C and that of the discarded blood samples.
MATERIALS AND METHODS
Area of study: Al-Ummah Medical Diagnostic Laboratory is in the capital city of Katsina State. This city known as Katsina has a population of about 432,194 making it one of the biggest cities in Katsina State. It operates in the WAT time zone. It is located around 12.99 latitudes and 7.60 longitudes and it is situated at elevation 519 meters above sea level. Al-Ummah Medical Diagnostic Laboratory has a total number of 14 Staff, 8 medical personnel and 6 non-medical personnel. It welcomes an average number of 10 patients per day.
Sampling procedure: Patients who visit Al-Ummah Medical Diagnostic Laboratory in the morning hours were sampled for this study for 3 weeks. Patients were enrolled in this research by convenience sampling. The sample was collected from the Al-Ummah Medical Diagnostic Laboratory in Katsina State, Nigeria from 1st through 21st June, 2019. A total of ten patients comprising 4 females and 6 males were used for the study. Five miles of blood samples were collected via intravenous using a 5 mL syringe and fashioned into two precise categories, freshly preserved blood samples and carelessly discarded blood samples. Two miles of the blood samples were kept in the EDTA bottle and stored at 4°C in the refrigerator (preserved group) while the remaining 3 mL in a syringe was kept in a refuse dump of the Laboratory and served as a discarded group. Both preserved groups and the discarded group were kept for 21 days. The study was carried out at Department of Biochemistry Laboratory of Abubaka Tafawa Belowa University (ABU), Zaria, Kaduna State, Nigeria from July to August, 2019.
Extraction of nuclear DNA: The DNA was extracted from blood samples using the phenol-chloroform method as described by Sambrook and Russell14. Exactly 200 μg of each of the samples was taken into separate tubes, to each of the tubes was added an equal volume of regents, Phenol, chloroform and RNAse. Firstly, 400 μL of lysis buffer was added, followed by 10 μL of proteinase K to inactivate the protein present in the samples vortex and mixed thoroughly. After that, it was incubated at 65°C for 10 min. Exactly 400 μL phenol-chloroform was added to the tubes and mixed thoroughly by vortex for 15 sec. The mixture was then centrifuged at 13,000 rpm for 10 min to pellet insoluble debris. About 200 μL of the upper layer was Pipetted into a new test tube. Then, 400 μL of phenol-chloroform was added to the extracted upper layer, then vortex and centrifuged 12,000 rpm for 5 min. Afterward, the upper layer of the centrifuged samples was pipetted into new tubes. Exactly, 1000 μL of 7% ethanol and 40 μL of sodium acetate were added to new tubes and incubated overnight at <-20°C (for maximum DNA). The mixture was centrifuged for 15 min at 4°C and the ethanol was discarded by inversion. About 400 μL of 70% ethanol was added and inverted. Then centrifuge at 4°C at 13,000 rpm for 10 min. After that the ethanol was discarded and the mixture was dried to obtain pure DNA in pellet form.
DNA purification: Genomic DNA extracted from participants was purified using the following protocol which involved 5 steps as described by Sambrook and Russell14.
RBC lysis: There was incubation of the sample with a 15-19 mL Autopure RBC Lysis solution for 5 min at room temperature to lyse the red blood cells. The samples were then centrifuged at 30006 g for 2 min to pellet the white blood cells.
Cell lysis and protein precipitation: To disperse the white blood cell pellet, 1.67 mL Autopure Precipitation Solution were vigorously dispensed and then 5 mL Autopure Cell Lysis Solution was added to lyse the white blood cells. The samples were mixed vigorously to precipitate the proteins and then centrifuged at 30006 g for 2 min. Five milliliters of Autopure 100% Isopropanol were added to the DNA-containing solution.
DNA precipitation: The output tubes were gently rotated 50 times to precipitate the DNA and then the samples were centrifuged at 30006 g for 2 min to pellet the DNA.
DNA wash: A dispense of 5 mL Autopure 70% Ethanol was done followed by centrifugation of the samples at 30006 g for 1 min to pellet DNA.
DNA hydration: DNA was rehydrated with DNA Hydration Solution according to the required DNA concentration defined by the users.
DNA quantification: The quantification of DNA was done by nanodrop and spectrophotometric methods as described by Sambrook and Russell14. Two different methods of quantification were used for measurement of DNA quantity: PicoGreen dsDNA quantitation assay and NanoDrop ND-8000 8 sample spectrophotometer. The PicoGreen dsDNA Quantitation Reagent is an ultra-sensitive fluorescent nucleic acid stain for quantitating double-stranded DNA (dsDNA). DNA samples were pipetted to 96-well plates for DNA concentration measurement with PicoGreen dsDNA quantitation assay and kit (Molecular Probes, Inc., The Netherlands). The NanoDrop ND-8000 8 sample spectrophotometer is a fullspectrum (220-750 nm) instrument that measures 8 individual 1 mL samples.
Determination of gel electrophoregraph of isolated DNA: The quality of DNA was determined by agarose gel electrolysis. The DNA in TE buffer was placed in a water bath at 65°C for 2 hrs. Samples were removed at the end of each period, separated in 0.7% agarose gel and stained with ethidium bromide. DNA samples (250 μg to 2 mg) were loaded onto 0.8-1% agarose gels and submitted to electrophoresis in TAE 0.5_ (40 mM Tris-acetate, 1 mM EDTA) electrophoresis buffer. Gels were stained 30 min in 0.5 mg mL–1 ethidium bromide or 1/10,000 SybrGreen_I (Molecular Probes). Gels were photographed with a Visiomic digital Imaging apparatus. Protocol as described by Sambrook and Russell14 was used.
Data analysis: Data were analyzed using SPSS 21.0 version package. An independent t-test was used to compare differences in the concentrations and quality of nuclear DNA obtained from the preserved group and discarded group.
RESULTS
Results of DNA extraction: Table 1 showed that the mean DNA yields from the preserved blood were higher than that of the discarded blood samples with a mean difference of 93.357.
Table 2 showed that the quality of DNA isolated from the preserved blood was higher than that of the discarded blood samples with a mean difference of 0.374.
| Table 1: | DNA concentration isolated from preserved and discarded blood samples | |||
| Samples identification numbers | Preserved samples (μg mL–1) |
Discarded samples (μg mL–1) |
| 1 | 221.4 |
183.17 |
| 2 | 253.15 |
162.31 |
| 3 | 242.43 |
139.62 |
| 4 | 268.23 |
141.23 |
| 5 | 263.69 |
106.18 |
| 6 | 285.69 |
147.2 |
| 7 | 144.91 |
125.71 |
| 8 | 235.06 |
110.44 |
| 9 | 255.65 |
139.98 |
| 10 | 174.76 |
155.56 |
| Mean±SD | 234.50±43.76 |
141.14±23.25 |
| Mean difference | 93.357 |
|
| t-value | 5.96 |
5.96 |
| p-value (variances) | 1.28 |
|
| p-value (means) | <0.00 |
<0.00 |
| F-value (variances) | 2.55 |
|
| Values are expressed as Mean±SD, means of t and variances of samples are significant at p<0.05 | ||
|
| Table 2: | DNA quality isolated from preserved and discarded blood samples | |||
| Sample identification numbers | Preserved samples (μg mL–1) |
Discarded samples (μg mL–1) |
| 1 | 1.87 |
1.53 |
| 2 | 1.86 |
1.35 |
| 3 | 1.84 |
1.54 |
| 4 | 1.94 |
1.49 |
| 5 | 1.9 |
1.44 |
| 6 | 1.74 |
1.54 |
| 7 | 1.98 |
1.46 |
| 8 | 1.9 |
1.63 |
| 9 | 1.97 |
1.69 |
| 10 | 1.93 |
1.52 |
| Mean±SD | 1.89±0.07 |
1.52±0.10 |
| Mean difference | 0.374 |
|
| t-value (18) | 9.97 |
9.97 |
| p-value (variances) | 0.55 |
|
| p-value | <0.00 |
<0.00 |
| F-value (variances) | 0.36 |
|
| Values are expressed as Mean±SD, means of t and variances of samples are significant at p<0.05 | ||
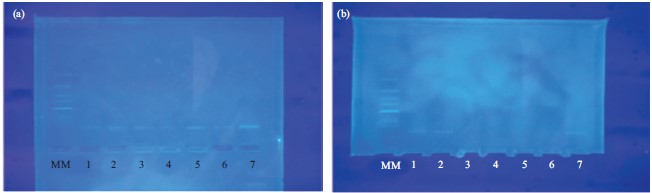
This showed that the gel electrophoresis of DNA isolated from preserved blood samples had a better hybridization pattern and no DNA degradation but in discarded blood, it had a poor hybridization pattern with severe DNA degradation. Both samples gave the same characteristic banding patterns (Fig. 1).
DISCUSSION
The DNA isolation and extraction has become a routine protocol for several analysis, either for diagnostic or experimental purpose. In each of this situation, a good concentration and quality is needed. Although, DNA is more stable than protein and is believed to be stable for a very long period, the source of DNA may also contribute to stability. In forensic science, the evidence is very important but for samples to be used particularly for tracing crime committed in the past or disputed paternity, biological samples play an important role in resolving the crisis. The degradation of DNA prolongs and complicates the resolution of a dispute.
From the result of the experiment in Table 1, the mean concentrations of DNA isolated from the preserved blood were significantly higher than that of the discarded blood samples, t (18) = 5.96, p = 0.00. The variances between the samples in both preserved and discarded blood samples showed no significant difference, F = 2.55, p = 1.28. The result of the study is in agreement with the report of Schwartz et al.15 and Hara et al.16. Thus, this implied that the variations in the concentrations of DNA in the investigated patients occurred by chance. The low mean concentrations of DNA isolated from the discarded blood samples might be as a result of bacterial nucleases dilapidation of the DNA.
In the same vein, Table 2 showed that the quality of DNA isolated from the preserved blood was significantly higher than the discarded blood samples with a mean difference of 0.374, t (18) = 9.97, p = 0.00. Also, there was a significant difference in the reported variances between the samples in both preserved and discarded blood samples, F = 0.36, p = 0.55. Thus, this indicated that the variations in the concentrations of DNA in the investigated patients occurred by chance. The DNA profiles obtained in the present study were concordant with the study of Ng et al.3, who reported that samples stored at the lower temperature exhibited significantly higher mean DNA yield. And that the decrease in DNA yield with elapsed time was also more gradual as compared to those stored at room temperature (Table 1 and 2). The work of Chen et al.17 and Hara et al.16 are in line with the result of this study.
In the same trend, the results of gel electrophoresis experiments are shown in Fig. 1. The DNA from preserved blood was more intact and most probably had higher molecular weight and less shearing than the DNA. These observed features could be attributed to less or no actions of bacterial nucleases dilapidation of the DNA. In the contrary, the discarded blood showed signs of severe degradation and such gave low molecular weight with poor hybridization pattern. This suggests that the preserved bloods are stable. The research Glasser et al.12, AlRokayan1 and Kim et al.18 are in accordance with the result of this present study. The implication of the study are that the information presented here is designed to facilitate informed decisions about the most appropriate sampling, preservation and extraction methodology relevant to the details of specific forensic cases to ensure sufficient DNA yield and successful analysis.
The isolation of high quantities of pure, intact, double stranded, highly concentrated, not contaminated genomic DNA is prerequisite for successful and reliable large-scale genotyping analysis. High quantities of pure DNA are also required for the creation of DNA-banks. The extraction of DNA is pivotal to forensic biochemistry. It is the starting point for numerous applications, ranging from fundamental research to routine diagnostic and therapeutic decision-making19. Extraction and purification are also indispensable in determining the unique characteristics of DNA, including its size, shape and function. The ability to extract DNA is of primary importance to study the genetic causes of disease and for the development of diagnostics and drugs. It is also vital for carrying out forensic science, sequencing of genomes, detecting bacteria and viruses in the environment and for determining paternity20.
From the results of the study, we recommend that samples meant for forensic applications and utilizations should be properly preserved and stored in the appropriate conditions. However, for plentiful and sufficient yields of DNA for other downstream analysis, the appropriate conditions should be maintained. We also recommend further PCR and chromatographic studies using larger sample size.
Although, there was a successful extraction of DNA from the both persevered and discarded groups, the sample size was a threat, the techniques employed in this study was not sufficient to establish a justifiable conclusion thus, techniques such as Qubit measurements, real-time PCR amplifications was lacking.
CONCLUSION
Based on the results of this study, it can be concluded that preservation has serious effects on the quantity and quality/stability of DNA. However, both samples are good for forensic investigations and molecular diagnosis of genetic diseases among others but preserved is better off.
SIGNIFICANCE STATEMENT
The findings highlight that preserved and discarded blood possessed an excellent DNA which can be beneficial as good tools for forensic investigations. This research finding is able to further support the growing interest in the employment of discarded blood in scientific exploration.
REFERENCES
- AlRokayan, S.A.H., 2000. Effect of storage temperature on the quality and quantity of DNA extracted from blood. Pak. J. Biol. Sci., 3: 392-394.
- Gill, P., A.J. Jeffreys and D.J. Werrett, 1985. Forensic application of DNA 'fingerprints'. Nature, 318: 577-579.
- Ng, H.H., H.C. Ang, S.Y. Hoe, M.L. Lim, H.E. Tai, R.C.H. Soh and C.K.C. Syn, 2018. Simple DNA extraction of urine samples: Effects of storage temperature and storage time. Forensic Sci. Int., 287: 36-39.
- Anchordoquy, T.J. and M.C. Molina, 2007. Preservation of DNA. Cell Preserv. Technol., 5: 180-188.
- Towne, B. and E.J. Devor, 1990. Effect of storage time and temperature on DNA extracted from whole blood samples. Hum. Biol., 62: 301-306.
- Dodd, B.E., 1985. Forensic science: DNA fingerprinting in matters of family and crime. Nature, 318: 506-507.
- Caboux, E., C. Lallemand, G. Ferro, B. Hémon and M. Mendy et al., 2012. Sources of pre-analytical variations in yield of DNA extracted from blood samples: Analysis of 50,000 DNA samples in epic. PLoS ONE, 7: e39821
- van der Hel, O.L., R.B. van der Luijt, H.B.B. de Mesquita, P.A.H. van Noord and B. Slothouber et al., 2002. Quality and quantity of DNA isolated from frozen urine in population-based research. Anal. Biochem., 304: 206-211.
- Shikama, K., 1965. Effect of freezing and thawing on the stability of double helix of DNA. Nature, 207: 529-530.
- Bulla, A., B. de Witt, W. Ammerlaan, F. Betsou and P. Lescuyer, 2016. Blood DNA yield but not integrity or methylation is impacted after long-term storage. Biopreserv. Biobanking, 14: 29-38.
- McCullough, J., B.J. Wieblen, P.K. Peterson and P.G. Quie, 1978. Effects of temperature on granulocyte preservation. Blood, 52: 301-310.
- Glasser, L., R.L. Fiederlein and D.W. Huestis, 1985. Liquid preservation of human neutrophils stored in synthetic media at 22 degrees C: Controlled observations on storage variables. Blood, 66: 267-272.
- Madisen, L., D.I. Hoar, C.D. Holroyd, M. Crisp, M.E. Hodes and J.F. Reynolds, 1987. The effects of storage of blood and isolated DNA on the integrity of DNA. Am. J. Med. Genet., 27: 379-390.
- Sambrook, J. and D.W. Russell, 2006. The Condensed Protocols from Molecular Cloning: A Laboratory Manual. 3rd Edn., Cold Spring Harbor Laboratory Press, United States, ISBN-13: 9780879697716, Pages: 800.
- Schwartz, A., A. Baidjoe, P.J. Rosenthal, G. Dorsey, T. Bousema and B. Greenhouse, 2015. The effect of storage and extraction methods on amplification of Plasmodium falciparum DNA from dried blood spots. Am. J. Trop. Med. Hyg., 92: 922-925.
- Hara, M., H. Nakanishi, K. Yoneyama, K. Saito and A. Takada, 2016. Effects of storage conditions on forensic examinations of blood samples and bloodstains stored for 20 years. Legal Med., 18: 81-84.
- Chen, W.C., R. Kerr, A. May, B. Ndlovu and A. Sobalisa et al., 2018. The integrity and yield of genomic DNA isolated from whole blood following long-term storage at -30°C. Biopreserv. Biobanking, 16: 106-113.
- Kim, Y.T., E.H. Choi, B.K. Son, E.H. Seo and E.K. Lee et al., 2011. Effects of storage buffer and temperature on the integrity of human DNA. Korean J. Clin. Lab. Sci., 44: 24-30.
- Ali, N., R. de Cássia Pontello Rampazzo, A.D.T. Costa and M.A. Krieger, 2017. Current nucleic acid extraction methods and their implications to point-of-care diagnostics. BioMed Res. Int., 2017: 9306564.
- Ghatak, S., R.B. Muthukumaran and S.K. Nachimuthu, 2013. A simple method of genomic DNA extraction from human samples for PCR-RFLP analysis. J. Biomol. Techn., 24: 224–231.
How to Cite this paper?
APA-7 Style
Nwankwo,
H., Uraku,
A., Orinya,
O., Enemchukwu,
B., Uraku,
O., Ezeani,
N., Obasi,
O., Nwankwo,
V. (2022). Effect of Storage on the Quantity and Quality of DNA Isolated from Preserved and Discarded Blood: Forensic Applications. Trends in Medical Research, 17(1), 1-7. https://doi.org/10.3923/tmr.2022.1.7
ACS Style
Nwankwo,
H.; Uraku,
A.; Orinya,
O.; Enemchukwu,
B.; Uraku,
O.; Ezeani,
N.; Obasi,
O.; Nwankwo,
V. Effect of Storage on the Quantity and Quality of DNA Isolated from Preserved and Discarded Blood: Forensic Applications. Trends Med. Res 2022, 17, 1-7. https://doi.org/10.3923/tmr.2022.1.7
AMA Style
Nwankwo
H, Uraku
A, Orinya
O, Enemchukwu
B, Uraku
O, Ezeani
N, Obasi
O, Nwankwo
V. Effect of Storage on the Quantity and Quality of DNA Isolated from Preserved and Discarded Blood: Forensic Applications. Trends in Medical Research. 2022; 17(1): 1-7. https://doi.org/10.3923/tmr.2022.1.7
Chicago/Turabian Style
Nwankwo, H.C., A.J. Uraku, O.F. Orinya, B.N. Enemchukwu, O.H. Uraku, N.N. Ezeani, O.U. Obasi, and V.O.U. Nwankwo.
2022. "Effect of Storage on the Quantity and Quality of DNA Isolated from Preserved and Discarded Blood: Forensic Applications" Trends in Medical Research 17, no. 1: 1-7. https://doi.org/10.3923/tmr.2022.1.7

This work is licensed under a Creative Commons Attribution 4.0 International License.